Please browse our services. If you are unsure about a service please contact us, and a MS&P professional will assist you.
See our services list below.
Additional information is listed, however if you require more detail please contact Dr. Arpad Somogyi for more information.
Proteomics:
- Cell Lysis
- Protein Quantitation
- Albumin Removal (MARS Column)
- 1D gel (mini gels 8 x 10 cm) and large format (20 x 24 cm)
- 2D gel (mini and large format gels)
- Multiplex stain (phosphorylation and glycosylation analyses)
- Protein ID – LC-MS/MS
- Post-Translation Modification Analysis
- MudPIT
- Quantitative Proteomics using Label Free Quantitation or iTRAQ
- Differential Gel Electrophoresis (DIGE)
- N-Terminal Sequencing
- LC-MALDI
- MALDI Imaging
- Western Blot
- IP Pulldowns
- OFFGEL Fractionation
Mass Spectrometry
- Accurate Mass Analysis to within 5 pmm (ESI and MALDI)
- Nominal Mass Analysis (ESI and MALDI)
- Structure determination
- LC-MS
- GC-MS
Metabolomics
We are extensively involved in both untargeted and targeted metabolomics studies and collaborations. Our high resolution instruments (Q-TOF and 15T FT-ICR) allow us to reliably determine chemical compositions of components of complex metabolomics mixtures. The targeted (or quantitative) metabolomics work is performed on our Quantiva QQQ instrument that has high sensitivity in the multiple reaction monitoring (MRM) mode. Part of the samples include those that are related to cancer and cardiovascular research.
Protein Expression and Purification
- Expression Optimization
- Large Scale Culture
- Protein Extraction and Purification
- Customer Provided Clone
- Generation and ID of Expression Positive Clones
- Subcloning Expression Vector
Proteomics
The Proteomics Shared Resource provides complete proteomic support from start to finish using the current state-of-the-art electrophoresis and imaging equipment, robotic sample handlers and mass spectrometers. See http://www.ccic.ohio-state.edu/MSP-Proteomics for details.
Accurate Mass

Accurate mass is used to determine the molecular weight of a sample to within 1 ppm on the Bruker 15 T FT-ICR instrument. It is used to verify a predicted molecular formula of a pure compound or of each individual components of complex mixtures (e.g., natural products, metabolomics mixtures, etc.). The analyte is mixed with an internal standard to use as a "lock mass" or internal calibration.
MS/MS

MS/MS or CID is used to determine structural or sequence information. The Esquire, and Q-TOF (ESI) can give fragment information. For ESI applications, the target molecule is isolated in the mass spectrometer and then collided with a gas to induce fragmentation. Due to the inherent nature of EI, molecules fragment as part of the ionization process. Here is an example of a tryptic peptide sequenced using the QTOF from Dr Micheal Freitas' research group. a
Polymer Distribution

Polymer distribution of polar and non-polar polymers can be determined using MALDI-TOF MS. Polymers must be soluble in water or organic solvent for analysis. Our software package on the MALDI allows for calculation of Mn, Mw.
Oligonucleotide Analysis

Oligonucleotides (DNA and RNA) can be analyzed by both MALDI and ESI. As with peptides and proteins, oligos are desalted prior to analysis, all buffers and salts must be removed for mass spec analysis as it suppresses ionization and many buffers are observed as background in the mass spectrum.
Molecular Weight Analysis

A simple molecular weight analysis can determine the presence or absence of a compound, purity, relative concentration and molecular weight. We can measure molecular weights as low as 50 Da and as high as 150,000 Da (and higher, no one has submitted anything bigger). ESI, EI and MALDI can all be used for this type of sample. Polymers, peptides, proteins, oligonucleotides are typically analyzed by simple molecular weight analysis. Most of our instruments are high resolution, so you can still get isotope information with the simple molecular weight analysis.
Protein/Peptide Analysis
Protein and peptides can be analyzed by both MALDI and ESI. Peptides and proteins are desalted prior to analysis, all buffers and salts must be removed for mass spec analysis as it suppresses ionization and many buffers are observed as background in the mass spectrum. As a guideline, we would need approximately 0.5 ug of a 10 KDa protein, 2.5 ug of a 50 KDa protein and 5 - 10 ug of a 100 KDa protein for good results. Please include the sequence if known at the time of sample submission.
GC-MS

Gas chromatography (GC) and mass spectrometry (MS) make an effective combination for chemical analysis. Among its uses are drug testing and environmental contaminant identification. The sample is injected through an injection port of the GC device, the GC instrument vaporizes the sample and then separates and analyzes the various components. Each component ideally produces a specific spectral peak and the time elapsed between injection and elution is called the "retention time." The size of the peaks is proportional to the quantity of the corresponding substances in the specimen analyzed. MS identifies the separated substances by using an electron impact ionization source (EI) which breaks the molecules into charged fragments and detected by the mass analyzer. A spectral plot displays the mass of each fragment under each peak in the GC plot. The compound can be identified by the GC retention time, the parent ion and fragmentation pattern as searched by a database of known compounds. The retention time can help to differentiate between some compounds.
LC/MS
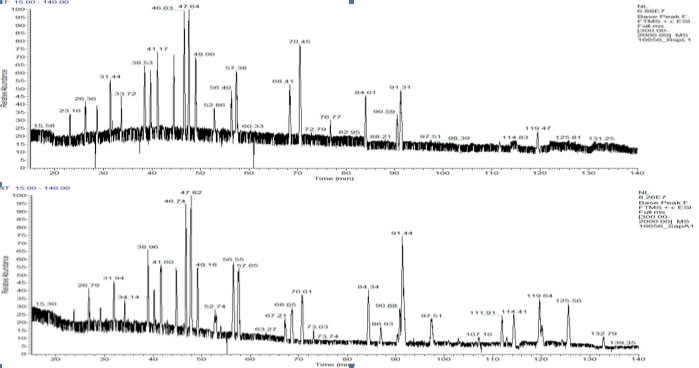
LC/MS is a very effective technique to combine peak detection with peak identification. Combining chromatography with mass spectrometry allows the chromatographer to "see inside" the chromatographic peak and to resolve co-eluting compounds of different molecular weights. Molecular weight information can identify predicted unknowns with better certainty and identify true unknowns by obtaining a "fingerprint' mass spectrum or fragmentation from CID and searching against commercial databases of spectra.



